{LivingNorwayR} (Chipperfield, Grainger, and Nilsen (2022)) is our newly developed R (R Core Team (2020)) package that allows the user to create a Darwin Core Standard (https://dwc.tdwg.org/) compliant data archive (“a data package”) for their biodiversity data.
We assume that the reader knows something about the Darwin Core Standards (a very basic understanding is okay). Have a look at our vignette “Handling Darwin Core Files With Living Norway: Example Using the TOV-E Dataset” for a good overview.
Here we will run through an example of how to map biodiversity data to the Darwin Core terms using {LivingNorwayR}.
Load the packages we need
As {LivingNorwayR} is still in development (although now firmly in the testing stage of development) we need to install it from GitHub. You can do this using the following code.
list.of.packages <- c("tidyverse", "devtools", "uuid")
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) install.packages(new.packages)
################################
devtools::install_github("LivingNorway/LivingNorwayR")
library(tidyverse, quietly = TRUE)
library(LivingNorwayR)
library(palmerpenguins)
The data
Let’s use a well-known and openly available dataset from R; The Palmer Penguins dataset (Horst, Hill, and Gorman (2020)).
This dataset consists of observations and measurements of three different species of penguin in Antarctica [Artwork by Allison Horst].
 We can have a quick look at the
dataset.
We can have a quick look at the
dataset.
penguin_data<-palmerpenguins::penguins_raw
#clean the column names
penguin_data<-penguin_data %>%
janitor::clean_names()
penguin_data %>%
head() %>%
kableExtra::kable() %>%
kableExtra::kable_styling("striped", full_width = F) %>%
kableExtra::scroll_box(width = "800px", height = "200px")
| study\_name | sample\_number | species | region | island | stage | individual\_id | clutch\_completion | date\_egg | culmen\_length\_mm | culmen\_depth\_mm | flipper\_length\_mm | body\_mass\_g | sex | delta\_15\_n\_o\_oo | delta\_13\_c\_o\_oo | comments |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAL0708 | 1 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A1 | Yes | 2007-11-11 | 39.1 | 18.7 | 181 | 3750 | MALE | NA | NA | Not enough blood for isotopes. |
| PAL0708 | 2 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N1A2 | Yes | 2007-11-11 | 39.5 | 17.4 | 186 | 3800 | FEMALE | 8.94956 | -24.69454 | NA |
| PAL0708 | 3 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A1 | Yes | 2007-11-16 | 40.3 | 18.0 | 195 | 3250 | FEMALE | 8.36821 | -25.33302 | NA |
| PAL0708 | 4 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N2A2 | Yes | 2007-11-16 | NA | NA | NA | NA | NA | NA | NA | Adult not sampled. |
| PAL0708 | 5 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N3A1 | Yes | 2007-11-16 | 36.7 | 19.3 | 193 | 3450 | FEMALE | 8.76651 | -25.32426 | NA |
| PAL0708 | 6 | Adelie Penguin (Pygoscelis adeliae) | Anvers | Torgersen | Adult, 1 Egg Stage | N3A2 | Yes | 2007-11-16 | 39.3 | 20.6 | 190 | 3650 | MALE | 8.66496 | -25.29805 | NA |
Each row is a single individual penguin of one of the three species. There are measures of body size (bill and flipper lengths), sex (male or female), as well as information on egg laying date and stable isotope analysis from blood samples.
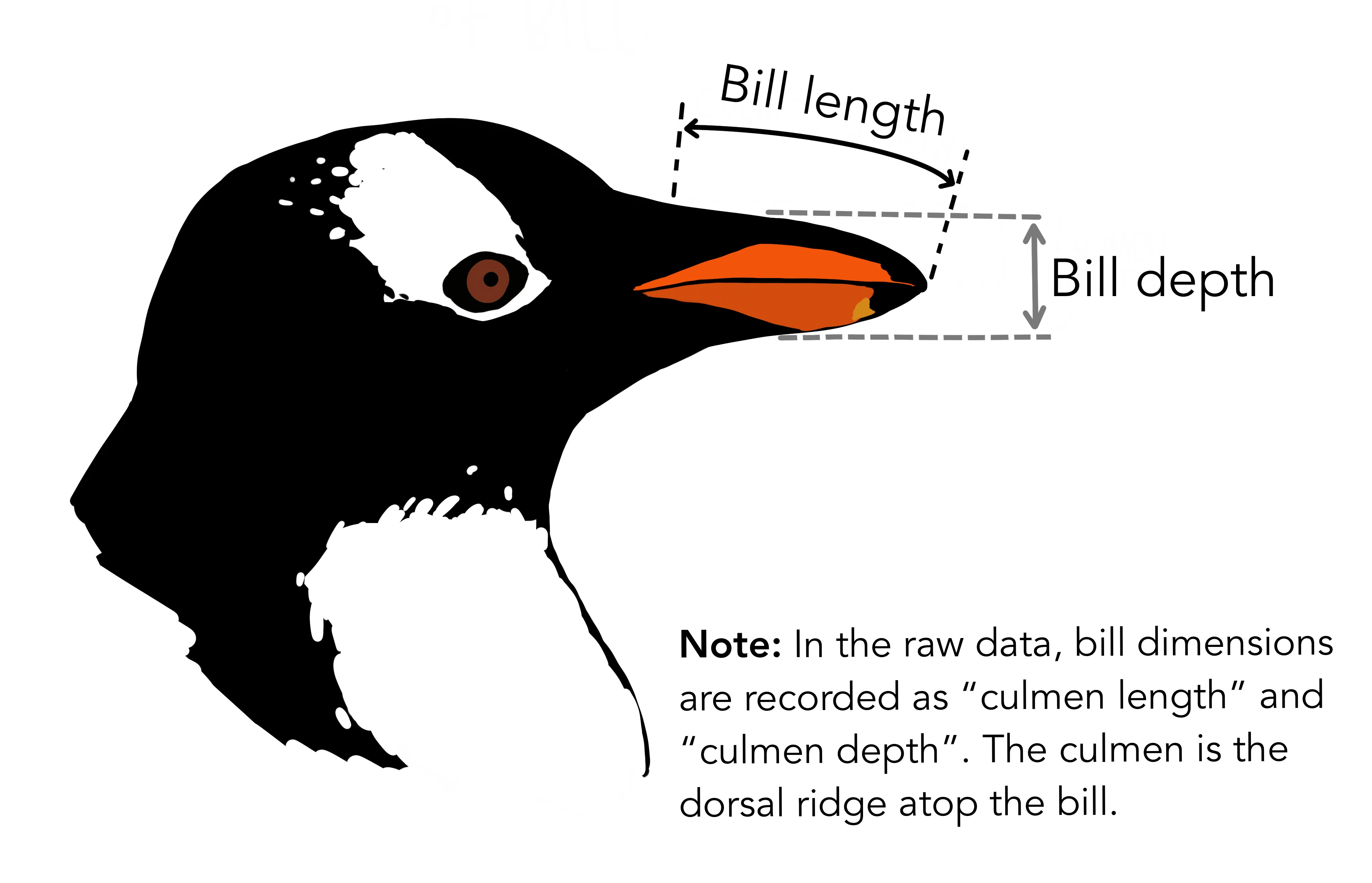
Mapping to Darwin Core
Deciding on the Core
The first task is to decide how our data will be structured. We need to decide what class of file will be our core data file in the Darwin Core Archive. {LivingNorwayR} can give us a list of the potential core classes that could best describe our data.
LivingNorwayR::getGBIFCoreClasses()
## $GBIFEvent
## http://rs.tdwg.org/dwc/terms/Event - Event
## An action that occurs at some location during some time.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/Event
## Version IRI: http://rs.tdwg.org/dwc/terms/version/Event-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## A specimen collection process. A camera trap image capture. A marine trawl.
## Miscellaneous information:
## DataSets/DataSet/Units/Unit/Gathering
##
## $GBIFOccurrence
## http://rs.tdwg.org/dwc/terms/Occurrence - Occurrence
## An existence of an Organism (sensu http://rs.tdwg.org/dwc/terms/Organism) at a particular place at a particular time.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/Occurrence
## Version IRI: http://rs.tdwg.org/dwc/terms/version/Occurrence-2020-08-20
## Type: Class
## Date modified: 2020-08-20
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## A wolf pack on the shore of Kluane Lake in 1988. A virus in a plant leaf in the New York Botanical Garden at 15:29 on 2014-10-23. A fungus in Central Park in the summer of 1929.
## Miscellaneous information:
## DataSets/DataSet/Units/Unit
##
## $GBIFTaxon
## http://rs.tdwg.org/dwc/terms/Taxon - Taxon
## A group of organisms (sensu http://purl.obolibrary.org/obo/OBI_0100026) considered by taxonomists to form a homogeneous unit.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/Taxon
## Version IRI: http://rs.tdwg.org/dwc/terms/version/Taxon-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## The genus Truncorotaloides as published by Brönnimann et al. in 1953 in the Journal of Paleontology Vol. 27(6) p. 817-820.
## Miscellaneous information:
## no simple equivalent in ABCD
The choice of which table should be the core table depends on how much information the data contains and also how the data owner wishes to share the data. There can be several different ways to structure the Darwin Core Archive that are still valid.
As each of the rows represents a sampling event we can use the Event table as our core.
Deciding on the extensions
We have species data for each Event and we can include an Occurrence table as an extension.
We can get a list of the other extension classes that are supported by GBIF using the following code:
LivingNorwayR::getGBIFExtensionClasses()[1:6] # Truncated
## $GBIFMultimedia
## http://rs.tdwg.org/ac/terms/Multimedia - Multimedia
## The Audubon Core is a set of vocabularies designed to represent metadata for biodiversity multimedia resources and collections. These vocabularies aim to represent information that will help to determine whether a particular resource or collection will be fit for some particular biodiversity science application before acquiring the media. Among others, the vocabularies address such concerns as the management of the media and collections, descriptions of their content, their taxonomic, geographic, and temporal coverage, and the appropriate ways to retrieve, attribute and reproduce them.
##
## IRI: http://rs.tdwg.org/ac/terms/Multimedia
## Version IRI: http://rs.tdwg.org/ac/terms/Multimedia
## Type: class
## Date modified: 2015-03-19
## Notes:
## The Audubon Core is a set of vocabularies designed to represent metadata for biodiversity multimedia resources and collections. These vocabularies aim to represent information that will help to determine whether a particular resource or collection will be fit for some particular biodiversity science application before acquiring the media. Among others, the vocabularies address such concerns as the management of the media and collections, descriptions of their content, their taxonomic, geographic, and temporal coverage, and the appropriate ways to retrieve, attribute and reproduce them.
## Miscellaneous information:
## GBIF core/extension class
## Vocabulary URI:
## http://terms.tdwg.org/wiki/Audubon_Core_Term_List
##
## $GBIFChronometricAge
## http://rs.tdwg.org/chrono/terms/ChronometricAge - ChronometricAge
## Extension to Occurrence Core to capture chronometric age information to be used only in cases where the collecting event is not contemporaneous with the time when the dwc:Organism was alive in its context. Collection event information can be reported in dwc:eventDate. See also the normative term list document at https://tdwg.github.io/chrono/list/ and the human-friendly Quick Reference Guide at https://tdwg.github.io/chrono/terms/.
##
## IRI: http://rs.tdwg.org/chrono/terms/ChronometricAge
## Version IRI: http://rs.tdwg.org/chrono/terms/ChronometricAge
## Type: class
## Date modified: 2021-03-27
## Notes:
## Extension to Occurrence Core to capture chronometric age information to be used only in cases where the collecting event is not contemporaneous with the time when the dwc:Organism was alive in its context. Collection event information can be reported in dwc:eventDate. See also the normative term list document at https://tdwg.github.io/chrono/list/ and the human-friendly Quick Reference Guide at https://tdwg.github.io/chrono/terms/.
## Miscellaneous information:
## GBIF core/extension class
## Vocabulary URI:
## https://github.com/tdwg/chrono/blob/master/vocabulary/term_versions.csv
##
## $GBIFIdentification
## http://rs.tdwg.org/dwc/terms/Identification - Identification
## A taxonomic determination (e.g., the assignment to a taxon).
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/Identification
## Version IRI: http://rs.tdwg.org/dwc/terms/version/Identification-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## A subspecies determination of an organism.
## Miscellaneous information:
## DataSets/DataSet/Units/Unit/Identifications/Identification
##
## $GBIFMeasurementOrFact
## http://rs.tdwg.org/dwc/terms/MeasurementOrFact - Measurement or Fact
## A measurement of or fact about an rdfs:Resource (http://www.w3.org/2000/01/rdf-schema#Resource).
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/MeasurementOrFact
## Version IRI: http://rs.tdwg.org/dwc/terms/version/MeasurementOrFact-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Notes:
## Resources can be thought of as identifiable records or instances of classes and may include, but need not be limited to dwc:Occurrence, dwc:Organism, dwc:MaterialSample, dwc:Event, dwc:Location, dwc:GeologicalContext, dwc:Identification, or dwc:Taxon.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## The weight of an organism in grams. The number of placental scars. Surface water temperature in Celsius.
## Miscellaneous information:
## Datasets/Dataset/Units/Unit/MeasurementsOrFacts or DataSets/DataSet/Units/Unit/Gathering/SiteMeasurementsOrFacts
##
## $GBIFResourceRelationship
## http://rs.tdwg.org/dwc/terms/ResourceRelationship - Resource Relationship
## A relationship of one rdfs:Resource (http://www.w3.org/2000/01/rdf-schema#Resource) to another.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/ResourceRelationship
## Version IRI: http://rs.tdwg.org/dwc/terms/version/ResourceRelationship-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Notes:
## Resources can be thought of as identifiable records or instances of classes and may include, but need not be limited to dwc:Occurrence, dwc:Organism, dwc:MaterialSample, dwc:Event, dwc:Location, dwc:GeologicalContext, dwc:Identification, or dwc:Taxon.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## An instance of an Organism is the mother of another instance of an Organism. A uniquely identified Occurrence represents the same Occurrence as another uniquely identified Occurrence. A MaterialSample is a subsample of another MaterialSample.
## Miscellaneous information:
## DataSets/DataSet/Units/Unit/Associations
##
## $GBIFEOLMediaExtension
## http://www.eol.org/schema/transfer#/EOLMediaExtension - EOLMediaExtension
## This extension draws from Audubon Core, Dublin Core and others to gather information about text and multimedia. It was designed to contain all the metadata that is required to be indexed by the Encyclopedia of Life (EOL), but this extension is hopefully general enough to be useful to all text and media providers and consumers. The original extension was offline; this is a copy recovered from the Internet Archive. The issue date is estimated.
##
## IRI: http://eol.org/schema/media/Document
## Version IRI: http://eol.org/schema/media/Document
## Type: class
## Date modified: 2010-01-01
## Notes:
## This extension draws from Audubon Core, Dublin Core and others to gather information about text and multimedia. It was designed to contain all the metadata that is required to be indexed by the Encyclopedia of Life (EOL), but this extension is hopefully general enough to be useful to all text and media providers and consumers. The original extension was offline; this is a copy recovered from the Internet Archive. The issue date is estimated.
## Miscellaneous information:
## GBIF core/extension class
## Vocabulary URI:
## http://eol.org/info/cp_archives
From this list the other extension class we could consider including in our Darwin Core Archive is the Measurement Or Fact table. This will hold information about the measurements taken.
The Event Core
We can get a list of the terms associated with an Event by using this code:
getGBIFEventMembers()[1:6] # Truncated
## $`http://purl.org/dc/terms/type`
## http://purl.org/dc/terms/type - Type (DEPRECATED)
## The nature or genre of the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/type
## Type: Property
## Date modified: 2008-01-14
## Is replaced by: http://purl.org/dc/elements/1.1/type
## Notes:
## To provide a string literal value for type, use dc:type rather than this term. In accordance with the Darwin Core RDF guide, rdf:type should be used instead of this term to indicate an IRI value for type.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2009-12-07_1
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## http://rs.tdwg.org/decisions/decision-2019-12-01_20
## Miscellaneous information:
## This term is deprecated and should no longer be used.
## not in ABCD
##
## $`http://purl.org/dc/terms/modified`
## http://purl.org/dc/terms/modified - Date Modified
## The most recent date-time on which the resource was changed.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/modified
## Version IRI: http://dublincore.org/usage/terms/history/#modified-003
## Type: Property
## Date modified: 2020-08-12
## Notes:
## Recommended best practice is to use a date that conforms to ISO 8601-1:2019.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## Examples:
## 1963-03-08T14:07-0600 (8 Mar 1963 at 2:07pm in the time zone six hours earlier than UTC). 2009-02-20T08:40Z (20 February 2009 8:40am UTC). 2018-08-29T15:19 (3:19pm local time on 29 August 2018). 1809-02-12 (some time during 12 February 1809). 1906-06 (some time in June 1906). 1971 (some time in the year 1971). 2007-03-01T13:00:00Z/2008-05-11T15:30:00Z (some time during the interval between 1 March 2007 1pm UTC and 11 May 2008 3:30pm UTC). 1900/1909 (some time during the interval between the beginning of the year 1900 and the end of the year 1909). 2007-11-13/15 (some time in the interval between 13 November 2007 and 15 November 2007).
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/language`
## http://purl.org/dc/terms/language - Language
## A language of the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/language
## Version IRI: http://dublincore.org/usage/terms/history/#languageT-001
## Type: Property
## Date modified: 2008-01-14
## Notes:
## Recommended best practice is to use an IRI from the Library of Congress ISO 639-2 scheme http://id.loc.gov/vocabulary/iso639-2
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/license`
## http://purl.org/dc/terms/license - License
## A legal document giving official permission to do something with the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/license
## Version IRI: http://dublincore.org/usage/terms/history/#license-002
## Type: Property
## Date modified: 2008-01-14
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-11-06_17
## Examples:
## http://creativecommons.org/publicdomain/zero/1.0/legalcode, http://creativecommons.org/licenses/by/4.0/legalcode
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/rightsHolder`
## http://purl.org/dc/terms/rightsHolder - Rights Holder
## A person or organization owning or managing rights over the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/rightsHolder
## Version IRI: http://dublincore.org/usage/terms/history/#rightsHolder-002
## Type: Property
## Date modified: 2008-01-14
## Examples:
## The Regents of the University of California
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/accessRights`
## http://purl.org/dc/terms/accessRights - Access Rights
## Information about who can access the resource or an indication of its security status.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/accessRights
## Version IRI: http://dublincore.org/usage/terms/history/#accessRights-002
## Type: Property
## Date modified: 2008-01-14
## Notes:
## Access Rights may include information regarding access or restrictions based on privacy, security, or other policies.
## Examples:
## not-for-profit use only, https://www.fieldmuseum.org/field-museum-natural-history-conditions-and-suggested-norms-use-collections-data-and-images
## Miscellaneous information:
## not in ABCD
From the above list of Event Members we can select those that are most relevant for our dataset. We do not have to use them all! GBIF recommends some required and suggested terms for Events here (https://www.gbif.org/data-quality-requirements-sampling-events). These include eventID, eventDate, samplingProtocol, samplingSizeValue and samplingSizeUnit as required. Some of the strongly recommended elements that it makes sense for us to include are parentEventID, countryCode, locationID decimalLatitude, decimalLongitude, geodeticDatum and coordinateUncertaintyInMeters. We can also add type,datasetName, ownerInstitutionCode, country, year, month and day.
Parent Events
Each event is a part of a higher level Event which is referred to as a Parent Event. The Parent Event in our case is represented by the “studyName” column. This represents a unique expedition carried out at a separate time. We can include this information in the Event table.
The Parent Events are three different expeditions in different years.
Each Parent Event needs a unique persistent identifier, parentEventID, which we can obtain from the {uuid} package (Urbanek and Ts’o (2021)).
penguin_data=penguin_data %>%
group_by(study_name) %>%
mutate(
parentEventID = uuid::UUIDgenerate(use.time = FALSE)
)
There are different date ranges for each parent event and these need to be added as an eventDate.
# Event Date for parentIDs
parent_penguinEvent=penguin_data %>%
group_by(parentEventID) %>%
summarise(min=min(date_egg), max=max(date_egg)) %>%
mutate(eventDate=paste0(min,"/", max)) %>% mutate(eventID=parentEventID)
We can also add some wider scale geographic information to the parent events. Such as continent and islandGroup.
# Event continent and islandGroup for parentIDs
parent_penguinEvent=parent_penguinEvent %>%
mutate(continent="Antarctica") %>%
mutate(islandGroup="Palmer Archipelago") %>%
select(!c(min,max))
Event
Let’s start with the type, datasetName and ownerInstitutionCode. The type is “Event”, the datasetName is “Palmer-penguins” and the Palmer Station Antarctica LTER ownerInstitutionCode is “PAL”.
penguin_data=penguin_data %>%
mutate(ownerInstitutionCode="PAL",
type="Event",
datasetName="Palmer-penguins")
Each Event also needs a unique identifier (eventID) and we can use the same approach as above. This time as each row is an Event we need to make sure the dataframe is ungrouped.
penguin_data=penguin_data %>%
ungroup() %>%
rowwise()%>%
mutate(
eventID = uuid::UUIDgenerate(use.time = FALSE)
)
Again we can include an eventDate for each event. For the penguins data we do not have a true date of the sampling event, but we do have a egg laying date and we shall use this for illustrative purposes. We can also extract the day, month and year information at the same time.
penguin_data=penguin_data %>%
mutate(
eventDate = date_egg) %>%
mutate(day=lubridate::day(date_egg),
month=lubridate::month(date_egg),
year=lubridate::year(date_egg))
As each event is a sample in the penguins dataset where they measured a individual penguin we can set the sampleSizeValue as 1. The sampleSizeUnit can be “Adult penguin”.
penguin_data=penguin_data %>%
mutate(sampleSizeValue=1) %>%
mutate(sampleSizeUnit="Adult penguin")
We can get the samplingProtocol for each event by looking at the original data package (links can be found when you type ??palmerpenguins::penguins_raw in to the R Console). All three parent events have the same protocol.
penguin_data=penguin_data %>%
mutate(samplingProtocol= "Each season, study nests, where pairs of adults were present, were individually marked and chosen before the onset of egg-laying, and consistently monitored. When study nests were found at the one-egg stage, both adults were captured to obtain blood samples used for molecular sexing and stable isotope analyses, and measurements of structural size and body mass. At the time of capture, each adult penguin was quickly blood sampled (~1 ml) from the brachial vein using a sterile 3 ml syringe and heparinized infusion needle. Collected blood was stored in 1.5 ml micro-centrifuge tubes that were kept cool. In the field, a small amount of whole blood was smeared on clean filter paper stored in a 1.5 ml micro-centrifuge tube for molecular sexing. Measurements of culmen length and depth (using dial calipers ± 0.1 mm), right flipper (using a ruler ± 1 mm), and body mass (using 5 kg ± 25 g or 10 kg ± 50 g Pesola spring scales and a weigh bag) were obtained to quantify body size variation. After handling, individuals at study nests were further monitored to ensure the pair reached clutch completion, i.e., two eggs. Molecular analyses were conducted at Simon Fraser University following standard PCR protocols, and stable isotope analyses were conducted at the Stable Isotope Facility at the University of California, Davis using an elemental analyzer interfaced with an isotope ratio mass spectrometer")
countryCode is the two letter standard (using ISO 3166-1-alpha-2) code for a country. Antarctica, defined as the territories south of 60°S is given the code AQ.
penguin_data =penguin_data %>%
mutate(countryCode="AQ",
country="Antarctica")
The locationID can be a global unique identifier or an identifier specific to the data set. We have the region and island for the penguins data so we can use these to develop a data specific identifier.
penguin_data=penguin_data %>%
mutate(locationID=paste0(region, "_", island))
We are not provided with precise coordinates (decimalLatitude, decimalLongitude) for the samples in the penguin data. However, we can get the centroid for each island; Torgersen is at -64.77308, -64.07413; Biscoe is at -65.4333316 -65.499998; and Dream is at -64.7333333, -64.2333333. The geodeticDatum is WGS 84 (this is the default assumed by GBIF if you do not add this field explicitly).
penguin_data=penguin_data %>%
mutate(decimalLatitude=
case_when(
island=="Torgersen"~-64.77308,
island=="Biscoe"~-65.4333316,
island=="Dream"~-64.7333333,
TRUE~as.numeric(NA))
) %>%
mutate(decimalLongitude=
case_when(
island=="Torgersen"~-64.07413,
island=="Biscoe"~-65.499998,
island=="Dream"~-64.2333333,
TRUE~as.numeric(NA))
) %>%
mutate(geodeticDatum="WGS 84")
As the coordinates are just the centroid for the island we need to include some measure of uncertainty (coordinateUncertaintyInMeters). We can guess the uncertainity by looking at the size of the islands. Torgersen is 400 m wide; Biscoe is around 500m wide and Dream is also around 400 m wide.
penguin_data=penguin_data %>%
mutate(coordinateUncertaintyInMeters=
case_when(
island=="Torgersen"~400,
island=="Biscoe"~500,
island=="Dream"~400,
TRUE~as.numeric(NA))
)
Finally, we can create the Event core by selecting those elements that we have listed above.
eventDF=penguin_data %>%
select(c("datasetName",
"ownerInstitutionCode",
"parentEventID",
"eventID",
"samplingProtocol",
"sampleSizeValue",
"sampleSizeUnit",
"eventDate",
"year",
"month",
"day",
"locationID",
"country",
"countryCode",
"decimalLatitude",
"decimalLongitude",
"geodeticDatum",
"coordinateUncertaintyInMeters"))
Then we need to add in the Parent Events in to the Event dataframe.
eventDF=eventDF %>%
mutate(continent=NA) %>%
mutate(islandGroup=NA) %>%
mutate(eventDate=as.character(eventDate)) %>% bind_rows(parent_penguinEvent)
The final stage is to initialise an event object in the {livingNorwayR} package - this will be used later to build the Darwin Core compliant data package.
GBIF_Event=initializeGBIFEvent(eventDF, idColumnInfo = "eventID", nameAutoMap = TRUE)
The Occurrence extension
We can find the supported terms for the Occurrence extension by using the following function.
getGBIFOccurrenceMembers()[1:6]#Truncated
## $`http://purl.org/dc/terms/type`
## http://purl.org/dc/terms/type - Type (DEPRECATED)
## The nature or genre of the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/type
## Type: Property
## Date modified: 2008-01-14
## Is replaced by: http://purl.org/dc/elements/1.1/type
## Notes:
## To provide a string literal value for type, use dc:type rather than this term. In accordance with the Darwin Core RDF guide, rdf:type should be used instead of this term to indicate an IRI value for type.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2009-12-07_1
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## http://rs.tdwg.org/decisions/decision-2019-12-01_20
## Miscellaneous information:
## This term is deprecated and should no longer be used.
## not in ABCD
##
## $`http://purl.org/dc/terms/modified`
## http://purl.org/dc/terms/modified - Date Modified
## The most recent date-time on which the resource was changed.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/modified
## Version IRI: http://dublincore.org/usage/terms/history/#modified-003
## Type: Property
## Date modified: 2020-08-12
## Notes:
## Recommended best practice is to use a date that conforms to ISO 8601-1:2019.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## Examples:
## 1963-03-08T14:07-0600 (8 Mar 1963 at 2:07pm in the time zone six hours earlier than UTC). 2009-02-20T08:40Z (20 February 2009 8:40am UTC). 2018-08-29T15:19 (3:19pm local time on 29 August 2018). 1809-02-12 (some time during 12 February 1809). 1906-06 (some time in June 1906). 1971 (some time in the year 1971). 2007-03-01T13:00:00Z/2008-05-11T15:30:00Z (some time during the interval between 1 March 2007 1pm UTC and 11 May 2008 3:30pm UTC). 1900/1909 (some time during the interval between the beginning of the year 1900 and the end of the year 1909). 2007-11-13/15 (some time in the interval between 13 November 2007 and 15 November 2007).
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/language`
## http://purl.org/dc/terms/language - Language
## A language of the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/language
## Version IRI: http://dublincore.org/usage/terms/history/#languageT-001
## Type: Property
## Date modified: 2008-01-14
## Notes:
## Recommended best practice is to use an IRI from the Library of Congress ISO 639-2 scheme http://id.loc.gov/vocabulary/iso639-2
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2019-12-01_19
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/license`
## http://purl.org/dc/terms/license - License
## A legal document giving official permission to do something with the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/license
## Version IRI: http://dublincore.org/usage/terms/history/#license-002
## Type: Property
## Date modified: 2008-01-14
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-11-06_17
## Examples:
## http://creativecommons.org/publicdomain/zero/1.0/legalcode, http://creativecommons.org/licenses/by/4.0/legalcode
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/rightsHolder`
## http://purl.org/dc/terms/rightsHolder - Rights Holder
## A person or organization owning or managing rights over the resource.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/rightsHolder
## Version IRI: http://dublincore.org/usage/terms/history/#rightsHolder-002
## Type: Property
## Date modified: 2008-01-14
## Examples:
## The Regents of the University of California
## Miscellaneous information:
## not in ABCD
##
## $`http://purl.org/dc/terms/accessRights`
## http://purl.org/dc/terms/accessRights - Access Rights
## Information about who can access the resource or an indication of its security status.
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://purl.org/dc/terms/accessRights
## Version IRI: http://dublincore.org/usage/terms/history/#accessRights-002
## Type: Property
## Date modified: 2008-01-14
## Notes:
## Access Rights may include information regarding access or restrictions based on privacy, security, or other policies.
## Examples:
## not-for-profit use only, https://www.fieldmuseum.org/field-museum-natural-history-conditions-and-suggested-norms-use-collections-data-and-images
## Miscellaneous information:
## not in ABCD
GBIF also has a list of required and recommended terms for Occurrence data (https://www.gbif.org/data-quality-requirements-occurrences). The required terms are occurrenceID, basisOfRecord,scientificName, eventDate (included in the event core). The recommended terms include countryCode, taxonRank,kingdom, decimalLatitude, decimalLongitude, geodeticDatum, coordinateUncertaintyInMeters,individualCount, organismQuantity and organismQuantityType, some of which are included in the event core.
We will also add some more information including type, collectionCode, organismQuantity, organismQuantityType, phylum, class, order, family, genus, vernacularName and sex.
The type is an “Occurrence”. The collectionCode can be “Palmer Station Antarctica LTER” and the occurrenceID should be a globally unique identifier.
penguin_data=penguin_data %>%
mutate(type="Occurrence",
collectionCode="Palmer Station Antarctica LTER") %>%
mutate(occurrenceID=uuid::UUIDgenerate(use.time = FALSE))
The basisOfRecord records how the observation was made (e.g. PreservedSpecimen, FossilSpecimen, LivingSpecimen, MaterialSample, Event, HumanObservation, MachineObservation, etc.)
klippy::klippy(position = "right")
penguin_data=penguin_data %>%
mutate(basisOfRecord="HumanObservation")
organismQuantity and organismQuantityType, are 1 individual penguin.
penguin_data=penguin_data %>%
mutate(organismQuantity= 1,
organismQuantityType= "individual")
For the scientificName and vernacularName we can use the species name from the original data set.
penguin_data=penguin_data %>%
mutate(scientificName=gsub("[\\(\\)]", "", regmatches(species, gregexpr("\\(.*?\\)",species))[[1]]),
vernacularName=gsub("\\s*\\([^\\)]+\\)","",species))
taxonRank, kingdom, phylum, class, order, family and genus all relate to the scientific name of the species. The taxonRank will be species because we have identified the penguins to the species level.
penguin_data=penguin_data %>%
mutate(kingdom="Animalia",
phylum="Chordata",
class="Aves",
order= "Sphenisciformes",
family= "Spheniscidae",
genus="Pygoscelis",
) %>%
mutate(taxonRank="species")
Finally, as we did with the Event core, we can create the occurrence extension by selecting those elements that we have listed above and create a {livingNorwayR} object.
occ_ext=penguin_data %>%
select(type,collectionCode,basisOfRecord,occurrenceID, organismQuantity, organismQuantityType,
eventID, eventDate, scientificName,kingdom, phylum, class, order, family, genus,vernacularName,taxonRank, sex)
GBIF_Occ=initializeGBIFOccurrence(occ_ext, idColumnInfo = "occurrenceID",nameAutoMap = TRUE )
The Measurement or Fact extension
Our final extension is the “Measurement or Fact extension”. We can look at the definition of this extension using the following code:
getGBIFExtensionClasses()$GBIFMeasurementOrFact
## http://rs.tdwg.org/dwc/terms/MeasurementOrFact - Measurement or Fact
## A measurement of or fact about an rdfs:Resource (http://www.w3.org/2000/01/rdf-schema#Resource).
##
## Defined in: https://dwc.tdwg.org/
## IRI: http://rs.tdwg.org/dwc/terms/MeasurementOrFact
## Version IRI: http://rs.tdwg.org/dwc/terms/version/MeasurementOrFact-2018-09-06
## Type: Class
## Date modified: 2018-09-06
## Notes:
## Resources can be thought of as identifiable records or instances of classes and may include, but need not be limited to dwc:Occurrence, dwc:Organism, dwc:MaterialSample, dwc:Event, dwc:Location, dwc:GeologicalContext, dwc:Identification, or dwc:Taxon.
## Executive committee decisions:
## http://rs.tdwg.org/decisions/decision-2014-10-26_15
## Examples:
## The weight of an organism in grams. The number of placental scars. Surface water temperature in Celsius.
## Miscellaneous information:
## Datasets/Dataset/Units/Unit/MeasurementsOrFacts or DataSets/DataSet/Units/Unit/Gathering/SiteMeasurementsOrFacts
GBIF has a list of properties for this extension (https://tools.gbif.org/dwca-validator/extension.do?id=dwc:MeasurementOrFact). These include measurementID measurementType, measurementValue, measurementAccuracy, measurementUnit, measurementDeterminedDate, measurementDeterminedBy, measurementMethod and measurementRemarks, none of which are required.
Let’s start with the measurementID. We should also include the occurrenceID and eventID so that we know which individual and in which sampling event the measurements were taken.
M_or_f=penguin_data %>%
mutate(measurementID=uuid::UUIDgenerate(use.time = FALSE))
There are a number of measurements that we can include in this extension.
M_or_f=M_or_f %>%
select(measurementID,parentEventID,occurrenceID,eventID,
culmen_length_mm, culmen_depth_mm, delta_13_c_o_oo, delta_15_n_o_oo, body_mass_g, flipper_length_mm)
We need to pivot the data so that all the measurement types go in to a single column called measurementType. All the measurements go in to a column called measurementValue.
M_or_f=M_or_f %>%
pivot_longer(cols = c(culmen_length_mm, culmen_depth_mm, delta_13_c_o_oo, delta_15_n_o_oo, body_mass_g, flipper_length_mm), names_to="measurementType",
values_to = "measurementValue"
) %>%
drop_na(measurementValue)
Finally we create a measurement or fact object using {livingNorwayR}.
GBIF_Measure=initializeGBIFMeasurementOrFact(M_or_f, idColumnInfo = "measurementID", nameAutoMap = TRUE)
Next steps
The next step is to write the metadata for the data. We can do this using the {LivingNorwayR} package and in the next tutorial we will show you how to do this (LINK TO PART TWO) and how to bring it all together in to a data package.
References
Chipperfield, Joseph, Matthew Grainger, and Erlend Nilsen. 2022. LivingNorwayR: Creates a Darwin Core Standard Compliant Data Archive (“a Data Package”) for Biodiversity Data. https://github.com/LivingNorway/LivingNorwayR.
Horst, Allison Marie, Alison Presmanes Hill, and Kristen B Gorman. 2020. Palmerpenguins: Palmer Archipelago (Antarctica) Penguin Data. https://allisonhorst.github.io/palmerpenguins/.
R Core Team. 2020. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
Urbanek, Simon, and Theodore Ts’o. 2021. Uuid: Tools for Generating and Handling of Uuids. https://CRAN.R-project.org/package=uuid.
